Table Of Contents
Previous topic
Next topic
This Page
Quick search
Enter search terms or a module, class or function name.
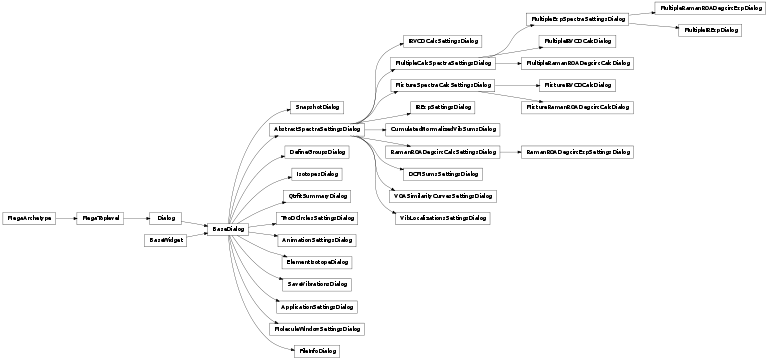
- BaseDialog
- base class for all dialogs
- MoleculeWindowSettingsDialog
- molecule window
- ApplicationSettingsDialog
- settings of PyVib2
- SaveVibrationsDialog
- dialog for saving vibrations
- IsotopesDialog
- customizing the isotopic composition
- ElementIsotopeDialog
- select an isotope for an element
- SnapshotDialog
- making a snapshot of a render widget
- DefineGroupsDialog
- defining groups in a molecule
- AnimationSettingsDialog
- dialog for saving animations
- AbstractSpectraSettingsDialog
- base class for settings of single mol
- RamanROADegcircCalcSettingsDialog
- Raman/ROA/Degcirc spectra
- IRVCDCalcSettingsDialog
- IR/VCD/g spectra
- MultipleCalcSpectraSettingsDialog
- base class for settings of sev. mols
- MultipleRamanROADegcircCalcDialog
- multiple Raman/ROA/Degcirc spectra
- MultipleIRVCDCalcDialog
- multiple IR/VCD/g spectra
- MultipleExpSpectraSettingsDialog
- base class for settings of sev. mols
- MultipleRamanROADegcircExpDialog
- multiple Raman/ROA/Degcirc exp. spectra
- MultipleIRExpDialog
- multiple infrared exp. spectra
- RamanROADegcircExpSettingsDialog
- experimental Raman/ROA/Degcirc spectra
- IRExpSettingsDialog
- experimental infrared absorption spectra
- MixtureSpectraCalcSettingsDialog
- mixture of spectra
- MixtureRamanROADegcircCalcDialog
- mixture of Raman/ROA/Degcirc spectra
- MixtureIRVCDCalcDialog
- mixture of IR/VCD/g spectra
- TwoDCirclesSettingsDialog
- settings of a circles canvas
- FileInfoDialog
- information about a file
- QtrfitSummaryDialog
- summary of a QTRFIT
- DCMSumsSettingsDialog
- sums of internuclear distances
- VOASimilarityCurvesSettingsDialog
- VOA similarity curves
- VibLocalizationsSettingsDialog
- localizations of vibrations
- CumulatedNormalizedVibSumsDialog
- cumulated normalized vibrational sums
- InvariantsContributionsDialog
- contributions of the Raman/ROA invariants to the intensities
| Author: | Maxim Fedorovsky |
|---|
Base class for all dialogs.
The _command() protected method is a handler for the button events. Subclasses should override it.
Initializer of the class.
| Parameter: | parent – parent widget |
|---|
Accepts the keyword arguments of Pmw.Dialog.
Settings of a molecule window.
Initializer of the class.
| Parameter: | molWindow – reference to the molecule window |
|---|
Settings of PyVib2.
Initializer of the class.
| Parameter: | mainApp – reference to the main window of PyVib2. |
|---|
Dialog for saving vibrations.
Initializer of the class.
| Parameter: | molWindow – reference to the molecule window |
|---|
Dialog for customizing the isotopic composition of a molecule.
Initializer of the class.
| Parameters: |
|
|---|
Dialog for selecting an isotope for an element.
Initializer of the class.
| Parameters: |
|
|---|
Dialog for making a snapshot of a render widget.
Initializer of the class.
| Parameters: |
|
|---|
Dialog for defining groups in a molecule.
Initializer of the class.
| Parameters: |
|
|---|
Dialog for saving animations of vibrations.
Initializer of the class.
| Parameter: | molWindow – reference to the molecule window |
|---|
Abstract class for settings of spectra.
Initializer of the class.
| Parameters: |
|
|---|
Select a page in the notebook.
Do nothing if the page does not exist.
| Parameter: | page – page name |
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
Settings of Raman/Roa/Degree of circularity spectra.
Initializer of the class.
| Parameters: |
|
|---|
Settings of IR/VCD/g spectra.
Initializer of the class.
| Parameters: |
|
|---|
Base class for settings of spectra for several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameters: |
|
|---|
Settings of Raman/Roa/Degree of circularity spectra of several mols.
Initializer of the class.
| Parameters: |
|
|---|
Settings of IR/VCD/g of circularity spectra of several mols.
Initializer of the class.
| Parameters: |
|
|---|
Base class for settings of exp. spectra for several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | figs (tuple or list) – figures to synchronize with |
|---|
Settings of Raman/Roa/Degree of circularity exp. spectra of sev. mols.
Initializer of the class.
| Parameters: |
|
|---|
Settings of infrared absorption exp. spectra of sev. mols.
Initializer of the class.
| Parameters: |
|
|---|
Settings of experimental Raman/Roa/Degree of circularity spectra.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
Settings of experimental infrared absorption spectra.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
Abstract class for settings of spectra of a mixture of molecules.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
Settings of Raman/ROA/Degree of circularity spectra of a mixture.
Initializer of the class.
| Parameters: |
|
|---|
Settings of IR/VCD/g of circularity spectra of a mixture.
Initializer of the class.
| Parameters: |
|
|---|
Settings of a circles canvas.
Initializer of the class.
| Parameters: |
|
|---|
Information about a file.
Initializer of the class.
| Parameters: |
|
|---|
Summary of a QTRFIT.
Initializer of the class.
| Parameters: |
|
|---|
Settings of the sums of internuclear distances.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
Settings of VOA similarity curves.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
Settings of a figure with localization of vibrations.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
Settings of a figure with cumulated normalized vibrational sums.
Initializer of the class.
| Parameters: |
|
|---|
Synchronize the GUI controls with a figure.
| Parameter: | fig – figure to synchronize with |
|---|
