Table Of Contents
Previous topic
Next topic
This Page
Quick search
Enter search terms or a module, class or function name.
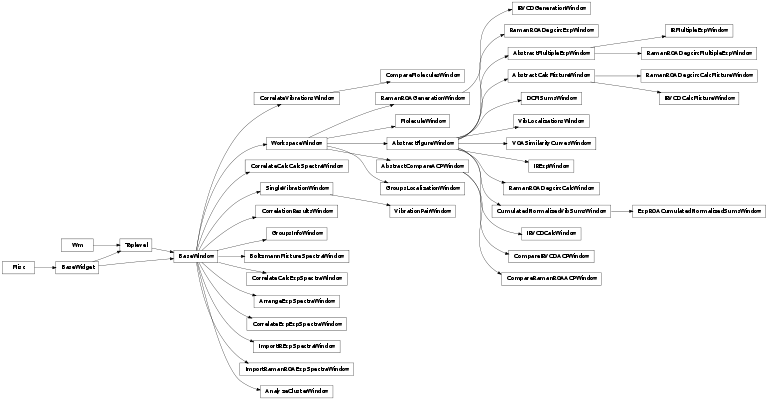
- BaseWindow
- base class of all windows
- WorkspaceWindow
- window that can be saved in a PyVib2 workspace
- MoleculeWindow
- exploring molecules
- CorrelateVibrationsWindow
- correlating vibrations
- CorrelationResultsWindow
- representing results of a correlation
- SingleVibrationWindow
- exploring a single vibration
- VibrationPairWindow
- exploring two vibrations simultaneously
- AbstractFigureWindow
- abstract class for windows with figures
- RamanROADegcircCalcWindow
- exploring Raman/ROA spectra
- RamanROAGenerationWindow
- Raman/ROA generation interface
- AbstractCalcMixtureWindow
- abstract class for mixture spectra
- RamanROADegcircCalcMixtureWindow
- exploring spectra of several molecules
- IRVCDCalcMixtureWindow
- exploring spectra of several molecules
- BoltzmannMixtureSpectraWindow
- spectra of a mixture of molecules
- IRVCDCalcWindow
- exploring IR/VCD spectra
- IRVCDGenerationWindow
- IR/VCD generation interface
- ImportRamanROAExpSpectraWindow
- importing exp. Raman/ROA spectra
- ImportIRExpSpectraWindow
- importing exp. infrared abs. spectra
- RamanROADegcircExpWindow
- exploring exp. Raman/ROA spectra
- IRExpWindow
- exploring exp. infrared abs. spectra
- DCMSumsWindow
- sums of internuclear distances
- ArrangeExpSpectraWindow
- select experimental spectra for one plot
- AbstractMultipleExpWindow
- abstract class for mult. exp. spectra
- RamanROADegcircMultipleExpWindow
- exploring sev. exp. Raman/ROA spectra
- IRMultipleExpWindow
- exploring sev. exp. infrared spectra
- CorrelateExpExpSpectraWindow
- correlating two experimental spectra
- CorrelateCalcExpSpectraWindow
- correlating calc. and exp. spectra
- CorrelateCalcCalcSpectraWindow
- correlating two calculated spectra
- CompareMoleculesWindow
- comparing molecule
- CompareRamanROAACPWindow
- comparing Raman/ROA ACPs
- CompareIRVCDACPWindow
- comparing IR/VCD ACPs
- AnalyzeClusterWindow
- analyzing clusters
- GroupsInfoWindow
- mark group(s) of atoms in a molecule
- GroupsLocalizationWindow
- show groups and localizations
- VOASimilarityCurvesWindow
- Raman/ROA similarity curves window
- VibLocalizationsWindow
- localizations of vibs on a fragment
- CumulatedNormalizedVibSumsWindow
- window with cumulated normalized vibrational sums for VROA
- ExpROACumulatedNormalizedSumsWindow
- window with experimental cumulated normalized sums of ROA
| Author: | Maxim Fedorovsky |
|---|
Base class for all windows of PyVib2.
The following public methods are exported:
- destroyme()
- destroy the window
- maximize()
- maximize the window
- clone()
- clone the window (should be overridden in subclases)
Initializer of the class.
| Parameters: |
|
|---|
Destroy the window.
| Parameter: | unregister – whether to unregister oneself from the Windows menu of the main window of PyVib2 |
|---|
Window that can be saved in a PyVib2 workspace.
Initializer of the class (kw are passed to BaseWindow).
Window for exploring molecules.
Initializer of the class.
| Parameters: |
|
|---|
Callback for the Apply/Ok buttons of the settings dialog.
| Parameter: | close_dialog – whether the dialog is to be closed |
|---|
Save an animation of the current vibration.
The gifsicle and ppm2fli programs must be installed.
| Parameters: |
|
|---|
Save vibrations.
| Parameters: |
|
|---|
All the keyword arguments must be supplied.
Window for correlating vibrations.
Initializer of the class.
| Parameters: |
|
|---|
Window for representing results of a correlation of vibrations.
Initializer of the class.
| Parameters: |
|
|---|
correlation_results must have the following keys:
start_time - formatted start time
t0 - start time as a tuple returned by os.times()
t1 - end time as a tuple returned by os.times()
ref_mol - reference molecule (pyviblib.molecule.Molecule)
tr_mol - trial molecule (pyviblib.molecule.Molecule)
- atom_pairs - atom_pairs of the fit fragment (null-based ndarray),
shape: (Natoms_F, 2) with Natoms_F being the number of atoms in the fragment atom numbers are one-based
- alignment - type of the orientationsl alignment
must be one of (‘Orientational’, ‘Inertia axes’,None)
include_tr_rot - if translations / rotations were included
- overlaps - overlaps matrix (one-based ndarray)
shape: (1 + NFreq_ref, 1 + NFreq_tr) with NFreq_ref and NFreq_tr being the number of the vibrations being correlated
- similarities - overlaps matrix (one-based ndarray),
shape: (1 + NFreq_ref, 1 + NFreq_tr)
camera - camera to be installed to the reference widget
if the orientational alignment has been performed:
rms - RMS value
q - quaternion (null-based ndarray), shape: (4,)
U - left rotation matrix (one-based ndarray), shape: (4, 4)
- ref_center - center of the reference molecule
(one-based ndarray), shape: (4,)
- fit_center - center of the fitted molecule
(one-based ndarray), shape: (4,)
| Parameters: |
|
|---|
Window for exploring a single vibration.
Initializer of the class.
| Parameters: |
|
|---|
Window for exploring two vibrations simultaneously.
No public methods are exported.
Initializer of the class.
| Parameters: |
|
|---|
Abstract class for widnows having spectra figures.
The canvas of the spectra is saved by resizing and there is a function for saving a figure.
Initializer of the class.
| Parameter: | mainApp – reference to the main window of PyVib2 |
|---|
Window for exploring Raman/ROA spectra.
Initializer of the class.
| Parameters: |
|
|---|
Abstract class for exploring spectra of several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Window for exploring Raman/ROA/Degree of circularity spectra of several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Window for exploring IR/VCD/g spectra of several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Raman/ROA generation interface.
Initializer of the class.
| Parameters: |
|
|---|
Window for exploring IR/VCD spectra.
Initializer of the class.
| Parameters: |
|
|---|
IR/VCD generation interface.
Initializer of the class.
| Parameters: |
|
|---|
Window for plotting spectra of a mixture of molecules.
Initializer of the class.
| Parameters: |
|
|---|
Window for importing experimetnal Raman/ROA spectra.
Initializer of the class.
| Parameter: | mainApp – reference to the main window of PyVib2 |
|---|
Window for importing experimental infrared absorption spectra.
Initializer of the class.
| Parameter: | mainApp – reference to the main window of PyVib2 |
|---|
Window for exploring experimental Raman/ROA/Degree of circularity spectra.
Initializer of the class.
| Parameters: |
|
|---|
Window for exploring experimental infrared absorption spectra.
Initializer of the class.
| Parameters: |
|
|---|
Representing the sums of internuclear distances.
Initializer of the class.
| Parameters: |
|
|---|
Arrange several experimental spectra to be put on one plot.
Initializer of the class.
| Parameters: |
|
|---|
Abstract class for exploring experimental spectra of several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Exploring experimental Raman/ROA/Degcree of circularity spectra of several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Exploring experimental infrared spectra of several molecules.
Initializer of the class.
| Parameters: |
|
|---|
Window for correlating two experimental spectra.
Initializer of the class.
| Parameters: |
|
|---|
Window for correlating a calculated and experimental spectrum.
Initializer of the class.
| Parameters: |
|
|---|
Window for correlating two calculated Raman/ROA or IR/VCD spectra.
Initializer of the class.
| Parameters: |
|
|---|
Window for comparing molecules.
One can superimpose the molecules, subtract ACPs if the data are available.
Initializer of the class.
| Parameters: |
|
|---|
Comparing Raman/ROA ACPs.
Initializer of the class.
| Parameters: |
|
|---|
Comparing IR/VCD ACPs.
Initializer of the class.
| Parameters: |
|
|---|
Analyzing clusters.
Initializer of the class.
| Parameters: |
|
|---|
Mark one or two groups of atoms in a molecule.
Initializer of the class.
| Parameters: |
|
|---|
Show available groups for a molecule and localizations on them.
Initializer of the class.
| Parameters: |
|
|---|
Raman/ROA similarity curves window.
Initializer of the class.
| Parameters: |
|
|---|
Representing the localizations of vibrations on a fragment.
Initializer of the class.
| Parameters: |
|
|---|
Window with cumulated normalized vibrational sums for VROA.
Initializer of the class.
| Parameters: |
|
|---|
Window with experimental cumulated normalized sums of ROA.
Initializer of the class.
| Parameters: |
|
|---|
